Effectiveness of vaccines against infection: living systematic review
Efficacy and effectiveness of COVID-19 vaccines against SARS-CoV-2 infection: interim results of a living systematic review, 1 January to 14 May 2021
We used an internal COVID-19 literature database constructed by the RKI library to search for relevant studies. This database covers PubMed, Embase (including Medline) and the preprint servers ArRvix, BioRxiv, ChemRxiv, MedRxiv, Preprints.org, ResearchSquare and Social Science Research Network (SSRN). We identified and screened a total of 4,870 entries. After screening 204 full-text articles, 30 studies were included. Two studies were RCTs, 19 studies had a cohort design and five were case–control studies, including two with test-negative design. One-dose efficacy/effectiveness was investigated in 24 studies (Table 1) and estimates ranged from 16.9% to 91.2%, with the majority of estimates ranging between 60% and 70%. In 17 of 26 studies, VE was reported after the second dose. Estimates ranged between 61.7% and 98.6%. These interim results of a living systematic review show that after completed course the EMA-approved COVID-19 vaccines have a VE of 80% to 90% in preventing SARS-CoV2 infections, including asymptomatic ones. At the time point of data cut for this interim analysis, only limited information was available on VOCs other than Alpha. Meanwhile, some studies have been published indicating reduced effectiveness against infections with VOC Delta for Comirnaty and Vaxzevria.
Safety, Immunogenicity, and Efficacy of the BNT162b2 Covid-19 Vaccine in Adolescents
Overall, 2260 adolescents 12 to 15 years of age received injections; 1131 received BNT162b2, and 1129 received placebo. BNT162b2 had a favorable safety and side-effect profile, with mainly transient mild-to-moderate reactogenicity (predominantly injection-site pain [in 79 to 86% of participants], fatigue [in 60 to 66%], and headache [in 55 to 65%]); there were no vaccine-related serious adverse events and few overall severe adverse events. Among participants without evidence of previous SARS-CoV-2 infection, no Covid-19 cases with an onset of 7 or more days after dose 2 were noted among BNT162b2 recipients, and 16 cases occurred among placebo recipients. The observed vaccine efficacy was 100% (95% CI, 75.3 to 100).
SARS-CoV-2 variant B.1.617 is resistant to bamlanivimab and evades antibodies induced by infection and vaccination
We analyze whether B.1.617 is more adept in entering cells and/or evades antibody responses. B.1.617 enters two of eight cell lines tested with roughly 50% increased efficiency and is equally inhibited by two entry inhibitors. In contrast, B.1.617 is resistant against bamlanivimab, an antibody used for COVID-19 treatment. B.1.617 evades antibodies induced by infection or vaccination, although less so than the B.1.351 variant. Collectively, our study reveals that antibody evasion of B.1.617 may contribute to the rapid spread of this variant.
Claim that Chinese team hid early SARS-CoV-2 sequences to stymie origin hunt sparks furor
[Science news report.] The unreviewed paper, by evolutionary biologist Jesse Bloom of the Fred Hutchinson Cancer Research Center, asserts that a team of Chinese researchers sampled viruses from some of the earliest COVID-19 patients in Wuhan, China, posted the viral sequences to a widely used U.S. database, and then a few months later had the genetic information removed to “obscure their existence.”
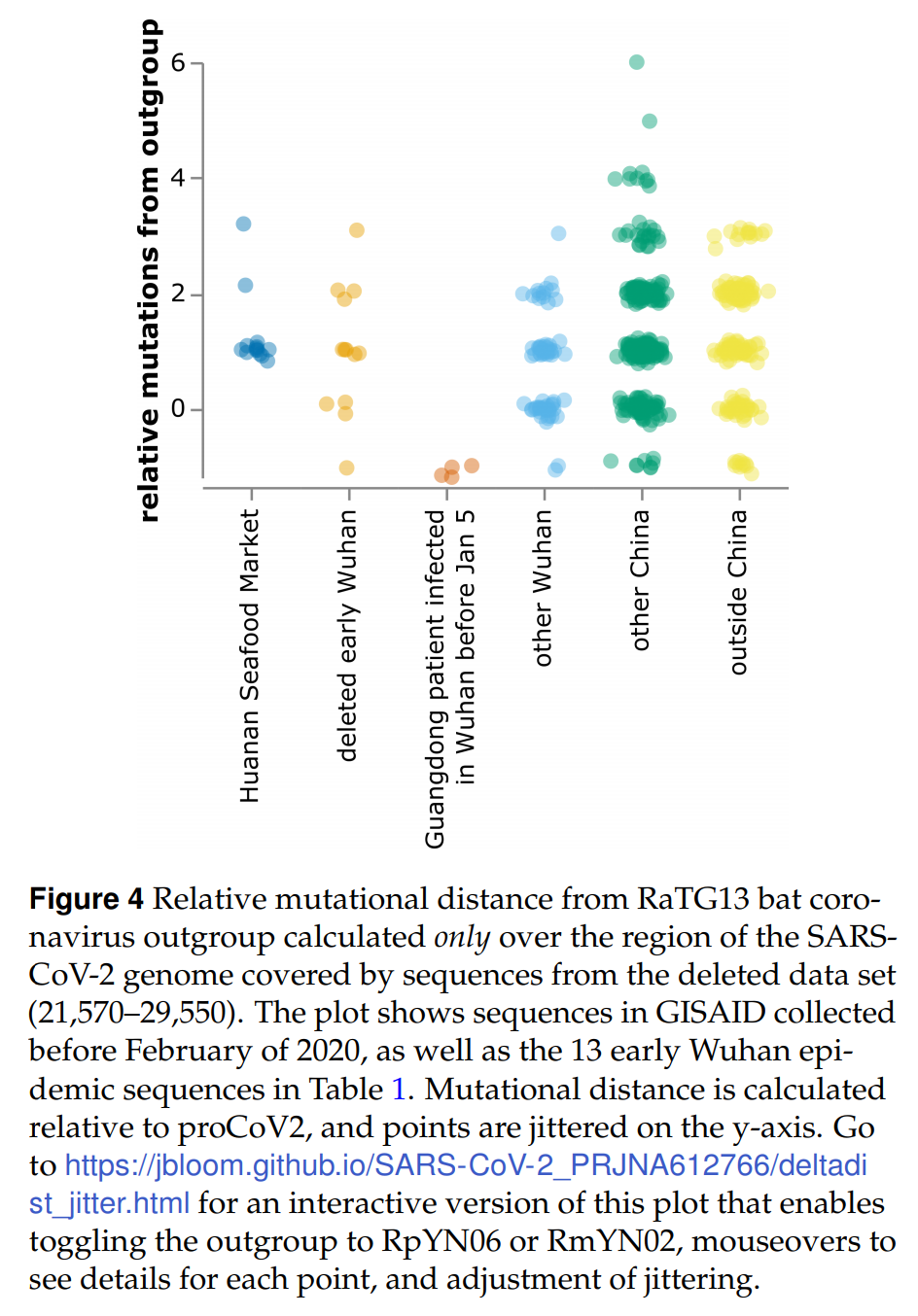
Recovery of deleted deep sequencing data sheds more light on the early Wuhan SARS-CoV-2 epidemic
Here I identify a data set containing SARS-CoV-2 sequences from early in the Wuhan epidemic that has been deleted from the NIH’s Sequence Read Archive. I recover the deleted files from the Google Cloud, and reconstruct partial sequences of 13 early epidemic viruses. Phylogenetic analysis of these sequences in the context of carefully annotated existing data further supports the idea that the Huanan Seafood Market sequences are not fully representative of the viruses in Wuhan early in the epidemic. Instead, the progenitor of currently known SARS-CoV-2 sequences likely contained three mutations relative to the market viruses that made it more similar to SARS-CoV-2’s bat coronavirus relatives.